Bioinformatics tools to advance virus diagnostics and epidemic tracing
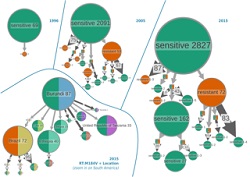
Recent advances in sequencing technologies include the development of NGS and bioinformatics-enabled rapid genetic analyses in individuals and populations. NGS analysis is becoming routine in research and clinical laboratories in the public and private sectors. However, despite the availability of millions of sequence fragments generated by NGS platforms, only a fraction was used until recently for virus discovery. The main obstacle is that current state-of-the-art methods in bioinformatics are too slow to rapidly and accurately classify and assemble known and unknown viruses using available NGS data from metagenomes. The main goal of the VIROGENESIS consortium was to develop new computational methods and software to overcome bioinformatics bottlenecks in the analysis and translation of NGS results for clinical and public health applications in the field of virology. Another important objective was the development of novel methods to characterise virome shifts in real-time during virus epidemics using large sets of NGS virus data. Finally, project partners aimed to provide user-friendly software that enables dynamic and interactive visualisation of the outbreak of epidemics in near real-time. Bioinformatics tools for virus discovery and clinical data analyses The VIROGENESIS consortium successfully developed algorithms and software applications for NGS data analyses overcoming major hurdles in the research of virus assembly and classification. This impressive development resulted in the creation of a fully functional web-based bioinformatics framework Genome Detective for quick identification of all known virus families using NGS and Sanger sequencing data. Increased flexibility and speed of the new applications enable analysis of subjects that include diagnostics, phylogeography, phylodynamics and transmission of drug resistance on a daily basis. VIROGENESIS tools accelerate the translation of NGS analysis results into investigation of microbial shifts, the source of transmission, the geographical origin and the extent of the spread of pathogens. Importantly, several final project developments for genomic analyses have been made available. These include desktop and multiplatform open-source software UGENE, which incorporates a suite of widely used bioinformatics tools. VIROGENESIS also collaborated with LUCIAD, the company that develops software for geospatial situational awareness solutions. This collaboration is helping to develop a platform for dynamic visualisations and the identification of ecological, demographic and epidemiological factors that impact virus spread. VIROGENESIS application in the current viral outbreaks During the recent Ebola epidemic, several international research teams were deployed to sequence Ebola virus for research and public health action. The VIROGENESIS software proved to be very efficient in several studies on recent outbreaks in Africa and South America. The discovery framework Genome Detective identifies viral pathogens directly from NGS data. Using these data together with additional VIROGENESIS tools enables informative visualisations and dynamic graphical user interfaces to follow epidemics in real time and space. An example of efficient detection of viral pathogens was presented in a study. They used VIROGENESIS tools and real-time sequencing data for genomic, spatial and epidemiological surveillance of the 2018 Yellow Fever Virus epidemic in Brazil. VIROGENESIS enables efficient analysis of NGS data for virus discovery, diagnostics and epidemic tracing. According to coordinators of the project: “With increased capture of genomic NGS data from virus epidemics and emerging outbreaks, we foresee an increased use of VIROGENESIS tools for analyses of these data, where rapid identification of virus type and transmission source is needed.”
Keywords
VIROGENESIS, NGS, bioinformatics, software, virus outbreaks, virus discovery