A fresh look at antibiotic design
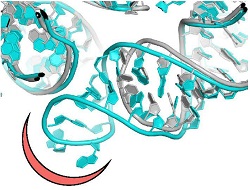
Most clinically useful antibiotics are natural compounds produced by microorganisms as defence against pathogens. To improve their potency, pharma companies chemically modify natural antibiotics or generate semi-synthetic analogues. However, pathogenic bacteria, such as Staphylococcus aureus, respond to the pressure imposed by natural and improved antibiotics, by developing resistance, significantly hampering their effectiveness. Intriguingly, antimicrobial resistance is a key survival tactic for many microorganisms and antibiotic resistance genes existed before the clinical utilisation of the natural antibiotics. Resistance manifests through various molecular mechanisms, such as activation of drug efflux pumps or mutations that modify the antibiotic binding pockets. Scientists of the EU-funded NOVRIB project focused their research on antibiotics that target ribosomes, multiprotein-RNA assemblies that drive protein biosynthesis. Given their key role in life, ribosomes constitute a target for many antibiotics, which in essence inhibit protein synthesis in pathogenic microorganisms. As Chemistry Nobelist and project coordinator Prof. Ada Yonath outlines “NOVRIB explored new potent selective compounds keeping in mind not only to combat or decrease antibiotics resistance but also to preserve the microbiome and the environment.″ Advancing ribosomal antibiotics Currently, antibiotic research mainly concentrates on underexplored microbial niches or the design of new chemical probes for improving the performance of existing antibiotics. The NOVRIB consortium proposed a different approach; they expanded the common search for establishing yet unknown mechanisms of antibiotic function, selectivity and resistance by discovering novel antibiotic binding sites. These sites demonstrated high potential to become useful targets for which resistance is less expected to appear soon. Towards this goal, they determined the detailed structure of ribosomes from the pathogenic bacteria, Staphylococcus aureus by X-ray crystallography or the recently developed 3D electron microscopy. By comparing this structure to those of ribosomes from non-pathogenic bacteria they identified unique structural motifs that provide novel sites for the design of new pathogen-specific drugs. “This paved the way for designing novel selective antibiotics, less susceptible to resistance, thus dealing with the current acute resistance issues,″ continues Prof Yonath. The future of antibiotics The emergence of multidrug-resistant strains, together with the very few new antibiotics that are presently in the drug discovery pipeline by pharma companies threaten to send us back to the pre-antibiotic era where infections were untreatable. Looking into the future, Prof Yonath is hopeful but emphasises “the immediate necessity for novel anti-bacterial agents.” Antibiotic toxicity – and the spreading of drug resistance – is linked to the chemical composition of many existing antibiotics which are non-biodegradable and indigestible by humans or animals. As a result, they contaminate the environment and, may enter agricultural irrigation systems, with direct implications for human and animal health. The novel antibiotics binding sites, which were discovered by this study, could also address this problem, since one can design according to preference compounds, with species specificity and minimal toxicity, thereby reducing the widespread antibiotics resistance while preserving the microbiome in an eco-friendly manner. In brief, contrary to the current medical preference of broad-spectrum antibiotics, the NOVRIB structural analysis approach provides the means for the design of pathogen-specific antibiotics that display selectivity, biodegradability and minimal toxicity. “Environmentally friendly drugs should also help reduce antibiotic resistance″ concludes Prof Yonath.
Keywords
NOVRIB, antibiotic, resistance, ribosome, bacteria