The unexpectedly complex issue of putting on and losing fat
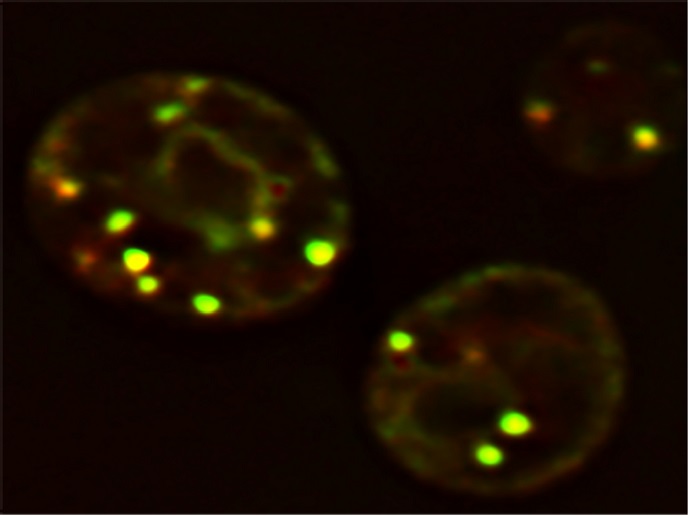
Understanding LD formation is central to uncover the pathophysiology of obesity and its associated diseases, like type 2 diabetes, atherosclerosis and fatty liver disease. Biogenesis of LDs occurs in the labyrinth of membranes in the cytoplasm, known as the endoplasmic reticulum (ER). See glowing yellow LDs in yeast cells in illustration above. Still, a mystery is what determines the ER sites of LD biogenesis. Supported by the Marie Skłodowska-Curie programme, “the main goal of the LD_Biogenesis project was to identify and characterise LD biogenesis sites in the ER,” explains the research fellow Vineet Choudhary.
Proteins collaborate with biogenesis factors
Research results demonstrated that LDs do not spontaneously form at random locations inside the ER, but originate at pre-defined discrete ER sites. Marked by the protein seipin, these collaborate with several LD biogenesis factors to establish droplet formation. What is interesting is that seipin is a non-enzymatic protein involved in lipodystrophy syndromes, a group of disorders characterised by selective loss of adipose tissue. “Seipin protein performs a decisive role in initiating LD biogenesis, lack of which results in LDs being born at ectopic sites,” says Choudhary. Results of the research have been published in the Journal of Cell Biology. Researchers used specially engineered baker’s yeast, Saccharomyces cerevisiae, to show where in the ER the LDs are assembled, and a novel probe that glows when hitting a site where triacylglycerol (TG) accumulates. With fluorescence and electron microscopy, the researchers mapped the relevant subdomains. By making genetic knock outs of LD biogenesis factors, the researchers showed that seipin together with the protein Nem1 are required to construct functional LD biogenesis sites. Seipin-Nem1 sites become associated with other LD biogenesis factors, including Pex30, a membrane-shaping protein, to facilitate birth and growth of LDs. Choudhary explains further the potential role of seipin. “We showed that if seipin is absent, it results in TG being synthesised at random locations in the ER, thereby resulting in improper LDs being assembled at wrong places.” How this irregular LD biogenesis manifests in lipodystrophy is an interesting question for further research.
Challenges and some surprise results
“We had to optimise several parameters to get the technique for correlative light and electron microscopy working,” explains Choudhary. “Similarly, we had to optimise expression and purification of enough TG synthase, sLro1, for performing in vitro lipid binding assays.” One of the perplexing findings of this study is the localisation of both seipin and Nem1 proteins at discrete ER sites, even in the absence of LDs. It will be interesting to know what drives their co-localisation at these ER subdomains. “Whether it is mediated by protein or lipid cues, or a combination of both remains to be determined. Knowing more about how these ER subdomains are formed will pave the way forward in deciphering the mechanism of LD biogenesis,” says Choudhary, enthusiastically, referring to his plans for future research.
Keywords
LD_Biogenesis, LD, ER, seipin, obesity, lipid droplet, endoplasmic reticulum