Software approach for MAPK understanding
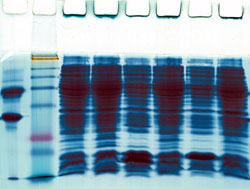
The EC-funded MAPK SIGNALLING project set out to elucidate the spatio-temporal organisation of the pathway and how that organisation impacts function. In other words, even though scientists are now aware of the pathway's set up, little is known regarding the pathway's involvement in all the different processes. Project partner Beatson Institute for Cancer Research focused on the development of the appropriate tools necessary for the processing of protein and gene expression data. These tools form an essential part of research efforts within the framework of the project, given the amount of information that needs to be processed and interpreted. Researchers tried to identify proteins that can be used for protein expression studies using 2-dimensional (2D) gels. The overall aim was to model the MAPK pathway using these high throughput proteomic data. Part of that goal was reached through the development of a specific portal used to manage 2D gel data and delineating protein-protein interactions. The online database was designed on the .NET platform and querying the database can be achieved through a variety of means. These include third party software and a set of standard queries built into the database.