Tackling drug resistance in the EU
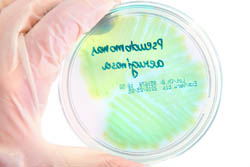
Europeans are becoming increasingly susceptible to bacterial infection, thanks in part to growing drug resistance. Innovative treatments are needed to fight these infections – especially those caused by superbug Gram-negative bacteria. The EU-funded project 'The identification, characterisation and exploitation of novel gram-negative drug targets' (AEROPATH) selected Pseudomonas aeruginosa (P. aeruginosa) bacteria as a model system and target. These bacteria cause around one tenth of the infections picked up during a hospital stay. They survive under a wide variety of conditions and are resistant to many antiseptics and all commonly used antibiotics. In addition, P. aeruginosa is highly adapted to develop resistance to antibiotics, making treatment even more difficult. Initially, AEROPATH examined, at the molecular level, the biology of P. aeruginosa. It identified routes via inhibition of high-value therapeutic targets, which weaken or interfere with the bacterium's ability to survive and cause infection. Next, over 35 selected proteins were genetically tested to investigate their suitability as drug targets. Some targets offered little potential regarding new drug development, which helped the AEROPATH team to effectively prioritise research on key proteins early on. This enabled the researchers not only to carry out the proposed project programme, but also to execute more experiments on high-value targets than was originally envisaged. As a result, the project has greatly enriched current scientific knowledge of P. aeruginosa drug targets, and has developed new tools for targeted antibacterial drug discovery. A range of mutant strains of P. aeruginosa and a mouse lung-infection model are now available for future research. There is also potential to extend the approach beyond the AEROPATH project and P. aeruginosa to other infectious diseases. The project database provides a template of how to best mine other pathogen genomes to rapidly identify and prioritise the most tractable targets. Furthermore, an efficient and cost-effective method for screening compound libraries against Gram-negative bacteria has been established.