Biosensor arrays act as a natural computer for detecting disease
Individual biosensors detect specific biological or chemical compounds to identify substances that may indicate health or environmental issues. But detecting multiple biomarkers using an array of biosensors can provide more comprehensive information for disease diagnosis or environmental monitoring. “The idea was to develop new types of computation using engineered bacteria,” explains project coordinator Christopher Barnes, professor in the Department of Cell and Developmental Biology at University College London, United Kingdom. These so-called biological computers process information from multiple inputs from engineered bacteria that use signalling molecules to communicate. “Using synthetic biology, also known as engineering biology, we put non-native genes into E. coli so that they can sense certain signals and send signals to each other,” he says. The genes were inserted on plasmids. “These are small pieces of DNA that we design on a computer and put into the cells. One cell type acts as a biosensor – it senses something in the environment and produces a communication signal.”
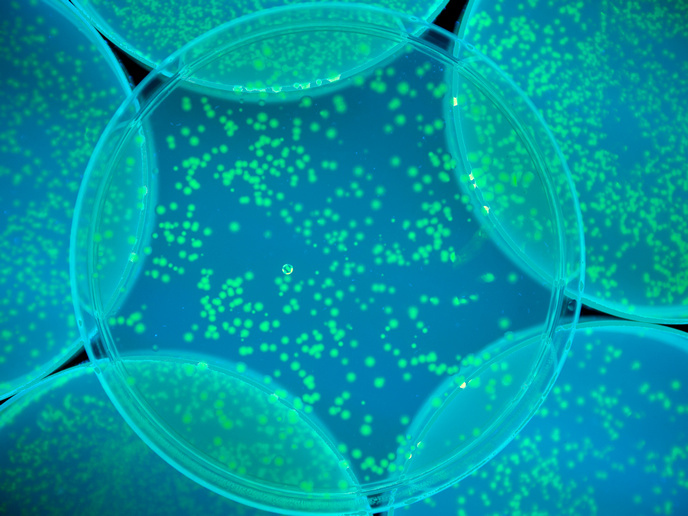
Cells that light up when they interact
Different cell types were engineered so that they responded to that signal and produced green fluorescent protein (GFP). “We could image them interacting,” Barnes says. “The cool thing about this is that we can place these engineered cells in different spatial orientations or patterns and they do different computations,” he adds, noting: “Certain colonies would turn on and off depending on how they were located next to each other and the signals in the environment.” “If certain combinations of input signals are present, the receiver cells express GFP. We measured the GFP response of these cells on the petri dish in the lab.”
Designing biosensors and arrays
Designing biosensors can be tricky. This was seen in the EU-funded DrugSense project for antibiotics sensors, and the CUSTOM-SENSE project that developed custom-made biosensors. But the SynBioBrain project, which was funded by the European Research Council, goes much further, designing whole arrays of biosensors. The team devised a mathematical model to design patterns of bacterial colonies that communicate with each other. “The complex processes involved in cell growth and nutrient diffusion were challenging to capture in mathematical models,” Barnes remarks, noting the partial differential equation model developed for the project also accounts for dynamic aspects, such as cell growth. Another problem was getting gene networks to respond with a high enough signal-to-noise ratio. “The approach we developed is quite sensitive to the positioning of the colonies. So we use a robot to position them,” he explains. These robots can be ‘noisy’, so further development would focus on better robots and reducing the number of bacteria in a colony to make the arrays smaller and faster.
Use of biosensors in diagnostics
The team used the computational technique to develop 10 biosensors in the lab, focusing on gut health and inflammation biomarkers. “The goal is to have an array of 20 biosensors that can do disease diagnostics. We’re not quite at that scale yet,” adds Barnes. Ultimately, devices can be developed where the engineered bacteria sit on a solid surface. Biological samples are then applied to it. “Instead of antibody-based testing, the device would recognise the [biosensor] signals from the bacteria directly,” he says. “If you had lots of these bacteria, all detecting different chemicals, you could build cheap biosensors. The bacteria can be freeze-dried on paper and used when needed.” Such diagnostic devices could be used at home or in challenging environments, without having to go to hospital.
Keywords
SynBioBrain, synthetic biology, biosensors, biomarkers, biological computers, E. coli, green fluorescent protein, DrugSense, CUSTOM-SENSE, gut health, diagnostic device